


Pol-MC was created by Dr Daniel Côté at the Ontario Cancer Institute (University of Toronto) with Dr Alex Vitkin and Brian Wilson. Funding provided by Natural Sciences and Engineering Research Council of Canada. For contact info, see below.
Summary
On this web page, you can download, compile and find instructions on how to use an implementation of a three-dimensional polarization-sensitive Monte Carlo algorithm for tissue optics. It has been introduced in an Optics Express article by Cote and Vitkin, and has undergone experimental validation (reflectance, transmittance, depolarization, optical rotation). It can be used for modelling the propagation of light (with or without polarization) in a collection of three-dimensional objects (with or without scatterers), with various sources (laser, isotropic). One can keep track of Stokes vector components, photons paths, interface crossing statistics, angle generation statistics, energy deposition, etc... It is programmed in C++ and is currently optimized for Unix (Mac OS X, Linux, etc...), although it should compile and run on Windows with very little modifications. It runs on clusters (i.e. you can combine the output of several runs together). There is no graphical user interface whatsoever and is not user-friendly. You will find more details (comments as well as some documentation) in the source code.
Polarimetry in tissue
To get an introduction to the code, please read: Daniel Cote and I. Alex Vitkin, "Robust concentration determination of optically active molecules in turbid media with validated three-dimensional polarization Monte Carlo model", Optics Express, 13, pp.148-163 (2005), available for free on the Optics Express web site.



|
| Fig.1 :Sample calculation showing the intensity I (left), Q (center) and U (right) Stokes parameters for a beam incident on the bottom of the cube of tissue with a scattering coefficient of 20 inverse centimeters. |


|
| Fig.2 :Sample calculation showing the intensity I (left), and the linear polarization Q (right) Stokes parameters for a beam incident on the bottom of the cube of tissue with a scattering coefficient of 0 to 60 inverse centimeters. The different frames represent different scattering coefficients. |


|
| Fig.3 : Sample calculation showing the energy deposited in a cube of tissue of scattering coefficient 20 inverse centimeter and absorption 1 inverse centimeter. On the left, the tissue does not contain any inclusion. On the right, there is a 0.3 x 0.3 x 0.3 cm inclusion with larger absorption coefficient (5 inverse centimeter) which absorbs most of the energy. |
|
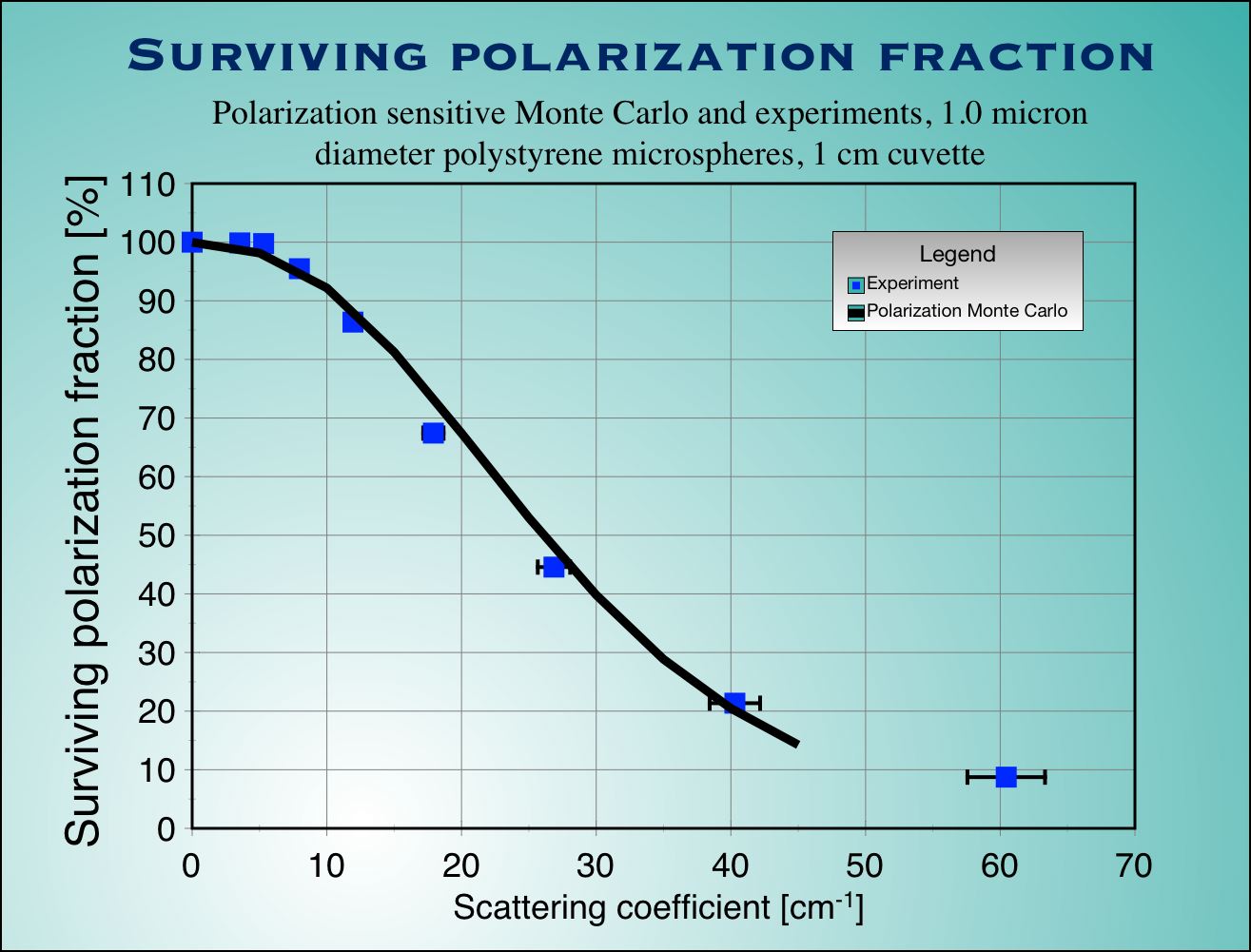
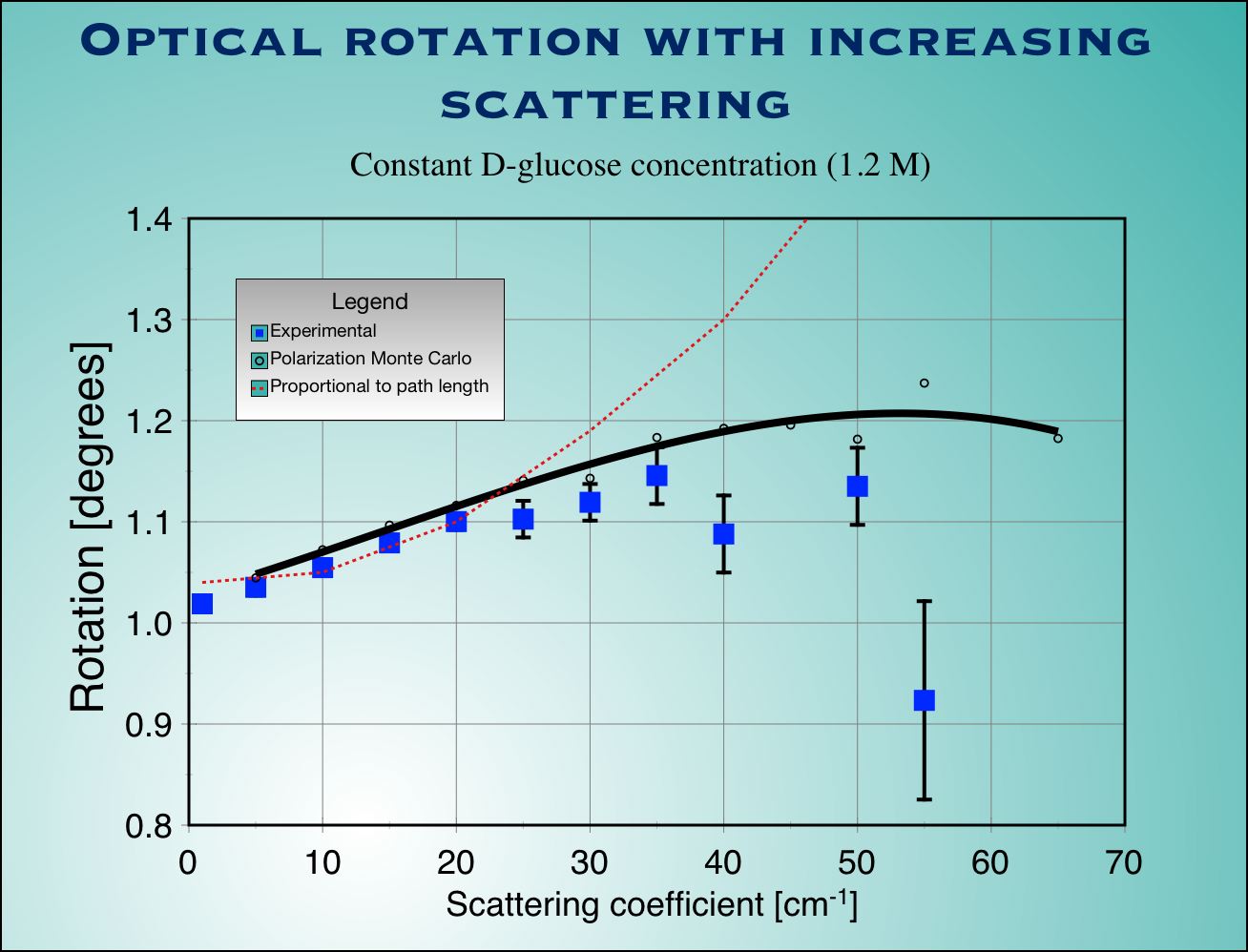
| Fig.4 : Experimental measurements (blue squaqres with error bars) and Monte Carlo calculation (not adjustable parameters used) for surviving polarization fraction (left) and optical rotation (right) as a function of scattering coefficient for a cubic sample of tissue. |
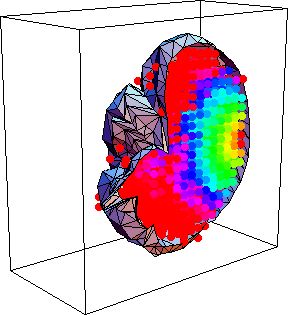 |
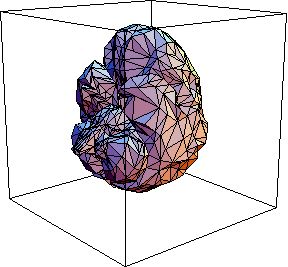
| Fig.5 : Three dimensional view of a brain and the energy deposited in it when illuminated from the top with a laser beam. The scattering coefficient is 200 per cm, calculation took 10 minutes. The few points outside the volume are a result of the discrete grid. |
Getting started
Foreword
The code available below has been tested to the extent that is described in the Optics Express article. More work is currently underway to test the rest of the algorithms and implementation. You shoould retrict your calculations to a single object, with or without polarization. Contact me for more info at dcote2@partners.org.
Download
First, you need the Pol-MC source code: polmc-2.5.1.tar.gz. Then you need the Thomas Germer's SCATMECH library for Mie scattering calculations:scatmech-3.0.tar.gz. Finally, it is convenient to have libxml2 (which is installed with most Linux distributions, but can also be downloaded from http://www.xmlsoft.org/).
Getting the program to work
Read the README file first, then read the parameter file (param.dat) then read the documentation folder to see a sample of the important classes (start with doc/MasterTOC.html). The main classes that do all the work are MCObject and Photon. The documentation is not complete: the source code is the ultimate guide. Instructions for Unix and Mac OS X are below. If you are on Windows, see the FAQ (below).
Without polarization support
In its most basic incarnation (i.e. no polarization needed), you only need polmc-2.5.1.tar.gz. On a unix workstation (on Mac OS X, use the Terminal), you would do the following:
With polarization support
Pol-MC was designed to consider polarization in calculations. The Mie scattering phase functions are calculated using SCATMECH. Currently, you need version 3.0 (altough version 4.0 is available on Thomas' web site). On a unix workstation, you would do the following:
This will install all the SCATMECH library files in your home directory's include and lib directory (and will create them if they don't exist). Now, you may compile polmc with polarization support. Download polmc-2.5.1.tar.gz, then do the following in the directory where polmc was downloaded:
Using polmc
Read the README file, included with the code. You use Pol-MC with: polmc -f polmc.log param.dat, where polmc.log is optional and param.dat is a parameter file similar in format to the param.dat file provided. The main program polmc is a simple demonstration program: it does not make use of all the features of the Pol-MC library. However, it makes use of the part that has been tested the most. Apart from the many parameters related to the scattering and optical properties, you will find the outputFilename parameter which determines the name of the output file. There is only output file, and its format is XML. XML stands for extended meta language format which is a text (i.e. human readable) format that looks like HTML. If you know how to use XML, XPath and Stylesheet transformations, it's extremely convenient and fast. If you don't, you can open the file in a text editor and copy/cut/paste the part you need. If you look at the output file (default is OutS1100.dat), you will find information on the objects, the parameters, the interface crossing statistics (I,Q,U,V, average path length, average number of collisions), the geometry (matlab patch, mathematica polygons, etc...).
Future work
There are scientific aspects and technical aspects to Pol-MC. Quite a few things may be added (dielectric fluctuations formalism, time-resolved photon paths, fluorecence to name a few). The first goal of Pol-MC is to be accurate, understandable, versatile, and fast (in that order). However, speed is always an issue with long calculations and Pol-MC could benefit from optimizations. The function names, parameters and others might change in the future.
FAQ
Where is the Graphical User Interface and how do I do simulate something more complicated than just a simple object?
There is no graphical user interface. Currently, you have two options: use simple forms (cube, ellispoid, single layer) with the "polmc" program compiled from main.cpp or brew your own by writing your own main() that constructs your own geometry (create object, put in world, create another object, place somewhere else, etc...). You may also contact me for help at dccote@novajo.ca.
I am on Windows. What do I do?
It is simple to compile Pol-MC on Windows, but I don't have access to a Windows PC with a compiler, hence I can't do it myself. Create a project and add all files ending with .h and .cpp. Then try to compile. If it works, great. If it does not, (this is very important), you need to edit config.h to reflect your compiler and define the preprocessor symbol HAVE_CONFIG_H. If you don't define HAVE_CONFIG_H, the config.h file does not get included (see for instance main.cpp).
Final word and contact info
Pol-MC is copyrighted by Daniel Côté, University Health Network and the Ontario Cancer Institute, University of Toronto. No warranties expressed or implied. You may use the code at your own risk and modify it. You are not bound to make your changes public, but you cannot sell this program for money. Any modifications that get sent to me to incorporate into the main code will be given proper credit. If you need information, I am now at CRISUMQ and my web site is www.dcclab.ca
